Jindřich Bär
(< Back to the list of posts) ( Collecting the gold-standard data for benchmarking )>
Benchmarking Charles Explorer search result rankings
This Python notebook shows the process of benchmarking the search result ranking for the Charles Explorer application. It is a part of my diploma thesis at the Faculty of Mathematics and Physics, Charles University, Prague.
Find out more about the thesis in the GitHub repository.
Made by Jindřich Bär, 2024.
In the thesis, we are trying to evaluate the benefits of using graph data metrics for reranking search results in an academic-purpose search engine. To carry out the evaluation, we need to establish a benchmarking process first. This notebook shows the process of benchmarking the search result ranking for the Charles Explorer application.
Creating the benchmark
Many search ranking metrics (e.g. DCG, nDCG etc.) require information about user-item interaction. This is used as a relevance signal to evaluate the quality of the ranking - if the ranking being evaluated ranks user-relevant items higher than irrelevant ones, it receives a higher score.
Unfortunately, the usage data from the Charles Explorer application are not statistically significant yet.
A proxy for an item relevance can be its position in other search engines’ results. By observing the result ranking for a given query in a different search engine, we can assign a relevance score to each item in the Charles Explorer search results. This relevance score can then be used to evaluate the quality of the ranking and can serve as a measure for the search engine’s performace, if we try to optimize it.
There are multiple academic search engines we can use as the “golden standard” for the benchmarking. Elsevier’s Scopus is one of the most popular academic search engines, with a large database of scientific articles in different fields. We can use it to benchmark the Charles Explorer search results, as Elsevier provides a free API for searching their database.
Other services (e.g. Web of Science, Google Scholar) were also considered, but they either do not provide a free API or have other limitations that make them unsuitable for this purpose - for example, with the Web of Science API, the results cannot be filtered by their affiliation to a particular institution. Since all the results in the Charles Explorer are from Charles University, this might cause at least some of the comparison results to be irrelevant.
Collecting the queries
As the first step, we need to sample the search query set for the benchmark. Since we want to rule out possible biases - or at least mitigate their impact - we need a large and diverse enough set of queries.
Generating these manually would be time-consuming and error-prone. It might also cause biases, as the queries would be generated by a single person, with a limited perspective, certain preferences and habits. To solve this issue, we can try to use a wordnet - a lexical language database (English, in our case). We can use it to generate a large set of diverse queries, perhaps less biased than a manually generated set.
We start by acquiring the synset for the lemma “field of study”. We then traverse the hyponyms (a word of more specific meaning) of this synset to get a broader set of words. By traversing this relation multiple times, we can search the space of the hyponyms “field of study” and generate a diverse set of queries.
Note: Preliminary experiments showed that the hyponyms of
field of studytend to underrepresent medical faculties in the search results. To mitigate this, we addmedicineas a second starting point for the traversal.
import wn
en = wn.Wordnet('oewn:2023')
ss = en.synsets('field of study') + en.synsets('medicine')
def expand_hyponyms(i: list[wn.Synset], depth: int = 1):
output = i
for step in range(depth):
new_hyponyms = []
for hypernym in output:
new_hyponyms += hypernym.hyponyms()
output += new_hyponyms
return output
queries = list(set([x.lemmas()[0] for x in expand_hyponyms(ss, 4)]))
queries
['statin',
'psephology',
'finance',
'pheniramine',
'topology',
'prosody',
'thiopental',
'NegGram',
'paediatrics',
'allergology',
'spectinomycin',
'Ponstel',
'Relanium',
'baby oil',
'specific',
'IP',
'sleeping capsule',
'gentian violet',
'descriptive anthropology',
'fluvastatin',
'physical science',
...
'chloramine',
'acoustics',
'pharmacokinetics',
'Pyridium',
'decongestant',
'meteorology',
'sedative',
'nontricyclic',
'Cox-2 inhibitor',
'soup',
'kagura',
'block',
'Mesantoin',
'asparaginase',
'physical anthropology',
'protriptyline',
'Pravachol',
'Relafen',
'fresco',
'thrombolytic',
'step dancing']
len(queries)
915
Using the process above, we managed to generate a set of 915 distinct queries - or query candidates.
From these, we want to create an unbiased set of queries for the benchmark with respect to the origin faculties of the results. To measure the faculty-wise bias, we can compare the distribution of the search results over faculties with the distribution of the total publication counts per faculty.
By querying the database, we can retrieve the total publication counts for each faculty and calculate the distribution of publication counts per faculty ($P_{\text{total}}$).
import pandas as pd
import numpy as np
import psycopg2
conn = psycopg2.connect("host=localhost port=5432 dbname=charles_explorer_db user=postgres password=charles_explorer_db_password")
cur = conn.cursor()
cur.execute('SELECT "A" as "faculty", COUNT(*) FROM "_FacultyToPublication" GROUP BY "A";')
records = cur.fetchall()
conn.close()
totals = pd.DataFrame(records, columns=[desc[0] for desc in cur.description])
totals['faculty'] = totals['faculty'].astype(int)
totals['prob'] = totals['count'] / totals['count'].sum()
totals.set_index('faculty', inplace=True)
totals['prob'].plot(kind='bar')
<Axes: xlabel='faculty'>
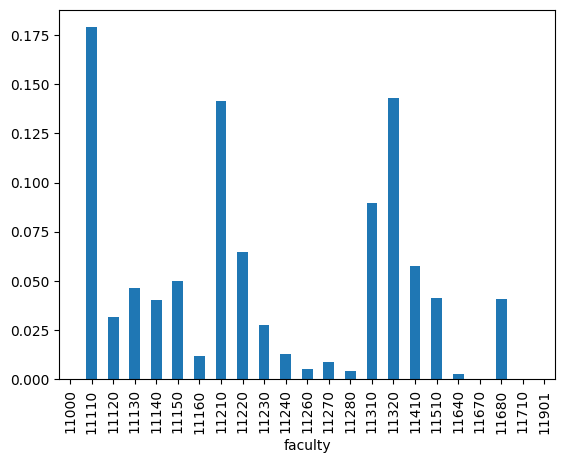
Now that we have the totals and sample queries “superset”, we can collect the search results from Charles Explorer.
We do this by querying the Charles Explorer API with each query candidate and storing the results as a row in Pandas DataFrame.
import pandas as pd
import requests
from collections import namedtuple
from urllib.parse import urljoin, urlencode, urlparse, urlunparse
def build_url(base_path: str, query: str):
# namedtuple to match the internal signature of urlunparse
Components = namedtuple(
typename='Components',
field_names=['scheme', 'netloc', 'url', 'path', 'query', 'fragment']
)
query_params = {
'query': query,
'_data': 'routes/$category/index'
}
return urlunparse(
Components(
scheme='http',
netloc='localhost:3000',
query=urlencode(query_params),
path='',
url=base_path,
fragment=''
)
)
def find_by_predicate(arr: list[dict], predicate):
return next((x for x in arr if predicate(x)), None)
def parse_response(response: requests.Response, query: str):
j = response.json()
query_ss = en.synsets(query)
return [
{
'id': result['id'],
'year': result['year'],
'name': find_by_predicate(result['names'], lambda x: x['lang'] == 'en').get('value') if find_by_predicate(result['names'], lambda x: x['lang'] == 'en') else result['names'][0].get('value'),
'faculty': len(result['faculties']) > 0 and result['faculties'][0].get('id') or -1,
'faculty_name': len(result['faculties']) > 0 and find_by_predicate(result['faculties'][0]['names'], lambda x: x['lang'] == 'en').get('value') or 'Unknown faculty',
'query': query,
} for result in j['searchResults']
]
def get_dataframe_for_queries(queries: list[str]):
df = pd.DataFrame()
for i, query in enumerate(list(set(queries))):
response = requests.get(build_url('/publication', query))
if i % 30 == 0:
print(f'Processed {i} queries')
# print(response.json())
df = pd.concat([df, pd.DataFrame(parse_response(response, query))])
return df
df = get_dataframe_for_queries(queries)
# Because of the big computational complexity, we store the query results in a file so we can reference them later without having to run the searches again.
df.to_csv('search_results.csv')
Processed 0 queries
Processed 30 queries
Processed 60 queries
Processed 90 queries
Processed 120 queries
Processed 150 queries
Processed 180 queries
Processed 210 queries
Processed 240 queries
Processed 270 queries
Processed 300 queries
Processed 330 queries
Processed 360 queries
Processed 390 queries
Processed 420 queries
Processed 450 queries
Processed 480 queries
Processed 510 queries
Processed 540 queries
Processed 570 queries
Processed 600 queries
Processed 630 queries
Processed 660 queries
Processed 690 queries
Processed 720 queries
Processed 750 queries
Processed 780 queries
Processed 810 queries
Processed 840 queries
Processed 870 queries
Processed 900 queries
df = pd.read_csv('./search_results.csv');
# we remove the publications that do not have a faculty associated with them - those don't contribute to the distribution of publications per faculty
df = df[~df['faculty'].isin([-1])]
df.reset_index().drop(columns=['index'], inplace=True)
df
| Unnamed: 0 | id | year | name | faculty | faculty_name | query | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 612484 | 2022.0 | Statins and liver | 11110 | First Faculty of Medicine | statin |
| 1 | 1 | 335691 | 2013.0 | Statins and myopathy | 11140 | Faculty of Medicine in Pilsen | statin |
| 2 | 2 | 594199 | 2016.0 | Do we use statins enough? News in the combinat... | 11140 | Faculty of Medicine in Pilsen | statin |
| 3 | 3 | 603996 | 2021.0 | Statins and COVID-19: Are they each other indi... | 11110 | First Faculty of Medicine | statin |
| 4 | 4 | 503698 | 2015.0 | Statins | 11110 | First Faculty of Medicine | statin |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 12509 | 25 | 287421 | 2014.0 | Within-Day and Weekly Variations of Thrombolys... | 11140 | Faculty of Medicine in Pilsen | thrombolytic |
| 12510 | 26 | 594262 | 2021.0 | Stroke 20 20: Implementation goals for intrave... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
| 12511 | 27 | 610284 | 2022.0 | Stroke thrombectomy (+- thrombolysis), and not... | 11120 | Third Faculty of Medicine | thrombolytic |
| 12512 | 28 | 479107 | 2011.0 | How accurately can the aetiology of cardiac ar... | 11680 | Central Library of Charles University | thrombolytic |
| 12513 | 29 | 113007 | 2010.0 | Endovascular Recanalization in the Treatment o... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
10167 rows × 7 columns
Having the actual search results, we can now compare its distribution to the distribution of total publication counts per faculty.
For this, we’ll be using KL divergence, a measure of how one probability distribution differs from a second, expected probability distribution. The value of KL divergence is always non-negative, with 0 meaning the two distributions are identical.
def get_subdf(df: pd.DataFrame, queries: list[str]):
return df[df['query'].isin(queries)]
def get_distribution(series: pd.Series):
return series / series.sum()
def get_series_for_queries(queries: list[str]):
return get_distribution(get_subdf(df, queries)['faculty'].value_counts().reindex(totals.index, fill_value=0)).fillna(0)
def get_kl_divergence(p: np.array, q: np.array):
return np.sum(p * np.log(p.add(1e-9) / q.add(1e-9)))
print(get_kl_divergence(get_series_for_queries(['modal logic']), totals['prob'])) # only returns results for one faculty (MFF)
print(get_kl_divergence(get_series_for_queries(queries), totals['prob']))
1.4029115694532561
0.04714136865559425
The example above shows two KL divergence computations.
The first computation compares the global faculty distribution with the distribution for the query “modal logic” - this query yields only results from the Faculty of Mathematics and Physics. The KL divergence is therefore high - the distribution of results is very different from the global distribution.
The second computation actually compares the faculty distribution on the ~900 sampled queries with the global distribution. The KL divergence is much lower, as the distribution of results is more similar to the global distribution.
Optimizing the KL divergence
Having defined the KL divergence as our metric, we can now reformulate the search for the unbiased set of queries as a minimization problem.
We want to find a subset of queries that minimizes the KL divergence between the distribution of search results and the distribution of total publication counts per faculty.
This problem is unfortunately not much different from the 0-1 Knapsack problem (or the subset sum problem), which are both NP-hard. Additionaly, we cannot simply reuse some of the existing algorithms for these problems, as many of those rely on the distributivity and associativity of the sum operation. This is however not the case for the KL divergence.
Similarly to the sum of the item values (in the Knapsack problem), the KL divergence is evaluated on the entire set, but unlike the sum, the items themselves do not have any “value” - and their contribution to the KL divergence changes depending on the other items in the set. This leaves us with a limited choice of algorithms to solve the problem. Because of the complexity of the problem and its smallish role in this work, we can use a simple random search. By sampling random subsets of a fixed size, we can try finding a subset that minimizes the KL divergence.
query_candidates = df['query'].unique()
def sample_noreplace(arr, n, k):
assert k <= len(arr)
idx = np.random.randint(len(arr) - np.arange(k), size=[n, k])
for i in range(k-1, 0, -1):
idx[:,i:] += idx[:,i:] >= idx[:,i-1,None]
return np.array(arr)[idx]
def get_best_queries(target_kl: float, max_epochs: int, n: int = 1000, k: int = 70):
best_queries = []
best_divergence = np.inf
for epoch in range(max_epochs):
print(f'Epoch {epoch}, best divergence: {best_divergence}')
query_groups = sample_noreplace(query_candidates, n, k)
divergences = [get_kl_divergence(get_series_for_queries(x), totals['prob']) for x in query_groups]
best_query_set = query_groups[np.argmin(divergences)]
divergence = divergences[np.argmin(divergences)]
if divergence < best_divergence:
best_queries = best_query_set
best_divergence = divergence
if divergence < target_kl:
break
return best_queries
best_query_set = get_best_queries(get_kl_divergence(get_series_for_queries(queries), totals['prob']) / 2, 10, 1000, 300)
Epoch 0, best divergence: inf
Epoch 1, best divergence: 0.03088109379075798
The randomized search has found a subset of queries that minimize the KL divergence to at least a half of the original value. This subset contains 300 queries, which we can use as the unbiased set of queries for the benchmark.
We can continue by plotting the distribution of search results and the distribution of total publication counts per faculty to see how well the search results represent the publication counts.
pd.DataFrame({
'totals': totals['prob'],
'all_queries': get_series_for_queries(best_query_set),
}).sort_values('totals', ascending=False).plot(kind='bar', figsize=(20, 10))
<Axes: xlabel='faculty'>
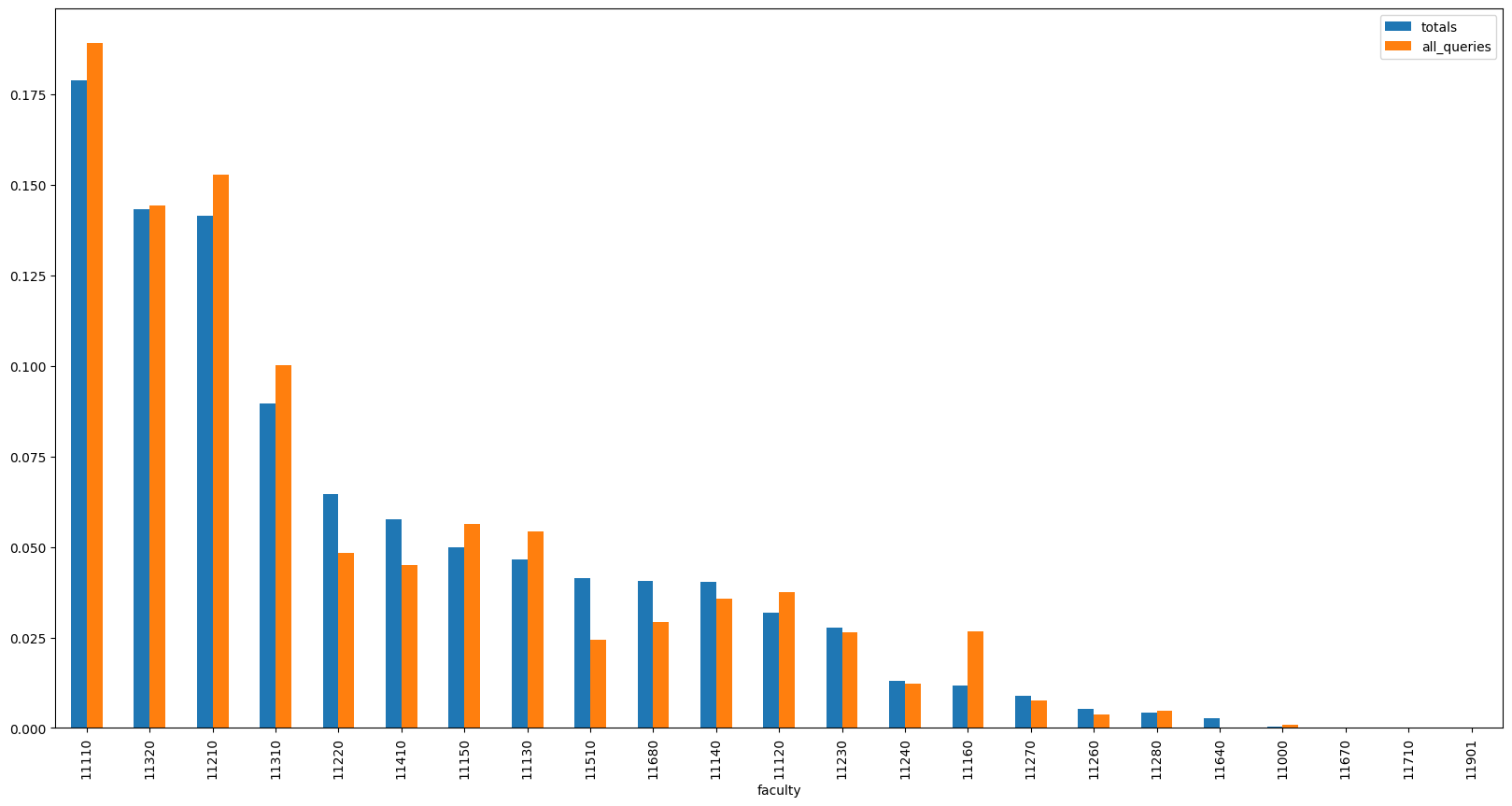
Even though the distributions are quite close to each other now, we can still see some of the faculties being overrepresented (and other being underrepresented) in the search results.
To further decrease the KL divergence, we can employ a simple hill climbing (or hill descending, in this case) algorithm. We can start with our random subset and iteratively try to remove a query from the subset. In each step of this process, we remove the query that, when removed, decreases the KL divergence the most. We repeat this process until the KL divergence stops decreasing.
Similarly to SGD, this method can get stuck in a local minimum. While this might cause the algorithm to not find the optimal solution, it should still be able to find a subset of queries with a low KL divergence.
current_divergence = get_kl_divergence(query_totals['prob'], totals['prob'])
new_divergence = 0
while True:
# try removing one query at a time, seeing which removal reduces the divergence the most
divergences = []
for query in best_query_set:
divergences.append(get_kl_divergence(get_series_for_queries([x for x in best_query_set if x != query]), totals['prob']))
divergences = np.array(divergences)
new_divergence = divergences.min()
if new_divergence < current_divergence:
# if current_divergence - new_divergence:
# print(f'No more queries to remove')
# break
print(f'Removing query {best_query_set[np.argmin(divergences)]} reduces divergence to {new_divergence}')
best_query_set = [x for i,x in enumerate(best_query_set) if i != np.argmin(divergences)]
current_divergence = new_divergence
else:
print(f'No more queries to remove')
break
Removing query penicillin reduces divergence to 0.016105491350444638
Removing query pas de trois reduces divergence to 0.015593487703697735
Removing query anti-inflammatory reduces divergence to 0.015155984041979853
Removing query alkylating agent reduces divergence to 0.014742787136988946
Removing query linguistics reduces divergence to 0.01437388632125047
Removing query vomit reduces divergence to 0.013995391804295524
Removing query photochemistry reduces divergence to 0.013631156512752046
Removing query expectorant reduces divergence to 0.013271099062614571
Removing query audiometry reduces divergence to 0.01295389196069935
Removing query sociolinguistics reduces divergence to 0.012613359059161467
Removing query antiprotozoal reduces divergence to 0.01230794480221695
Removing query linear programming reduces divergence to 0.012025689986302714
Removing query NSAID reduces divergence to 0.0117049979000795
Removing query sulfadiazine reduces divergence to 0.011442551618439099
Removing query frontier reduces divergence to 0.011166341603609633
Removing query hydroxychloroquine reduces divergence to 0.010869342262337331
Removing query freeze reduces divergence to 0.010617804931890167
Removing query history reduces divergence to 0.01037022471793504
Removing query antidiuretic reduces divergence to 0.010111961592201575
Removing query mechanical engineering reduces divergence to 0.009851994516792181
Removing query laxative reduces divergence to 0.009616712686632136
Removing query dermatology reduces divergence to 0.009400117125337445
Removing query genealogy reduces divergence to 0.009170328760585428
Removing query radiopharmaceutical reduces divergence to 0.00895568915479433
Removing query topology reduces divergence to 0.008722330414410707
Removing query psychodynamics reduces divergence to 0.008482114095765478
Removing query geochemistry reduces divergence to 0.008234283046945087
Removing query poetics reduces divergence to 0.008000773290104254
Removing query spasmolytic reduces divergence to 0.007785975076860612
Removing query bioscience reduces divergence to 0.007605583927328279
Removing query rhetoric reduces divergence to 0.00742463703721507
Removing query pharmacokinetics reduces divergence to 0.007235321971843382
Removing query high energy physics reduces divergence to 0.007003329499052894
Removing query immunology reduces divergence to 0.0068278835555687775
Removing query dialectic reduces divergence to 0.0066355710980884235
Removing query diltiazem reduces divergence to 0.0064749145945258825
Removing query capsule reduces divergence to 0.006332669209236989
Removing query variation reduces divergence to 0.0061831708138719126
Removing query menthol reduces divergence to 0.006057261977463686
Removing query econometrics reduces divergence to 0.005938469602979169
Removing query epidemiology reduces divergence to 0.005790835727612971
Removing query Oriental Studies reduces divergence to 0.0056524881017620725
Removing query beta blocker reduces divergence to 0.0055374731213476
Removing query civics reduces divergence to 0.0054193257071314515
Removing query taxonomy reduces divergence to 0.005312823010956788
Removing query otolaryngology reduces divergence to 0.0051872734103167485
Removing query classical philology reduces divergence to 0.005067817612199804
Removing query virology reduces divergence to 0.004973629393371306
Removing query set theory reduces divergence to 0.004868857811316672
Removing query nuclear medicine reduces divergence to 0.004781839591283388
Removing query natural science reduces divergence to 0.004700693563307107
Removing query Occidentalism reduces divergence to 0.0046300243421567066
Removing query SSRI reduces divergence to 0.004548713128497094
Removing query cosmography reduces divergence to 0.0044842917218509805
Removing query oxytetracycline reduces divergence to 0.0044199835921090586
Removing query barbiturate reduces divergence to 0.004368837971897515
Removing query object reduces divergence to 0.004308423062073141
Removing query alpha blocker reduces divergence to 0.0042450953249444255
Removing query oceanography reduces divergence to 0.004192541566260031
Removing query quantum physics reduces divergence to 0.004151420293851628
Removing query dose reduces divergence to 0.004098453004089903
Removing query phonemics reduces divergence to 0.004035013976505352
Removing query power engineering reduces divergence to 0.0039951033390309585
Removing query aeronautics reduces divergence to 0.003967212944591768
Removing query strategics reduces divergence to 0.003938690465785155
Removing query streptomycin reduces divergence to 0.0039054473602914037
Removing query ology reduces divergence to 0.0038749510271096337
Removing query English reduces divergence to 0.0038420646599258883
Removing query polymyxin reduces divergence to 0.0038097651743487864
Removing query computational linguistics reduces divergence to 0.003778289249139709
Removing query codeine reduces divergence to 0.0037391257265411474
Removing query cultural studies reduces divergence to 0.0037007293385864447
Removing query medicine reduces divergence to 0.003659366111778401
Removing query chemical engineering reduces divergence to 0.0036304184058123766
Removing query cryptography reduces divergence to 0.0036104536260615078
Removing query structural linguistics reduces divergence to 0.003589262499551347
Removing query atropine reduces divergence to 0.003550016448961152
Removing query Indocin reduces divergence to 0.0035348400120714467
Removing query alpha-interferon reduces divergence to 0.0035195748319500272
Removing query application reduces divergence to 0.0034988921641933455
Removing query complementary medicine reduces divergence to 0.0034750675271526337
Removing query hydraulic engineering reduces divergence to 0.0034566931778127487
Removing query antimetabolite reduces divergence to 0.0034360798569208297
Removing query disulfiram reduces divergence to 0.0034192066261994935
Removing query hermeneutics reduces divergence to 0.0034013913935241787
Removing query tubocurarine reduces divergence to 0.0033846736202896932
Removing query theory of probability reduces divergence to 0.003366610698795755
Removing query sedative-hypnotic reduces divergence to 0.0033509682412260376
Removing query dermatoglyphics reduces divergence to 0.0033384667678033842
Removing query human ecology reduces divergence to 0.0033245508498037195
Removing query sulfanilamide reduces divergence to 0.003311749513610583
Removing query orthodontia reduces divergence to 0.0033047967004089995
Removing query yellow jacket reduces divergence to 0.003297533665479544
Removing query behaviorism reduces divergence to 0.0032897518571533115
Removing query neuropsychiatry reduces divergence to 0.00326810919682544
Removing query castor oil reduces divergence to 0.0032514546415738244
Removing query chemical science reduces divergence to 0.003239460477727438
Removing query strategy reduces divergence to 0.003230062278393327
Removing query cephalothin reduces divergence to 0.003222187605007859
Removing query Paxil reduces divergence to 0.003217801027944129
Removing query hydromorphone reduces divergence to 0.003213321464772427
Removing query salol reduces divergence to 0.003208603977272919
Removing query patent medicine reduces divergence to 0.0032038695454998873
Removing query antimycin reduces divergence to 0.0031998468918678423
Removing query symbology reduces divergence to 0.0031946072720307602
Removing query sulindac reduces divergence to 0.003193170076465984
Removing query demulcent reduces divergence to 0.0031917975765188658
Removing query military science reduces divergence to 0.0031902106212587018
Removing query peace and conflict studies reduces divergence to 0.00318909131528973
Removing query ritual dance reduces divergence to 0.003187852837685122
Removing query biomedicine reduces divergence to 0.003185264035967492
Removing query landscape architecture reduces divergence to 0.0031813441642548675
Removing query Glucotrol reduces divergence to 0.00317780811369104
Removing query thioguanine reduces divergence to 0.003175405864741112
Removing query pure mathematics reduces divergence to 0.0031740782329779628
Removing query tyrothricin reduces divergence to 0.003173120688959345
Removing query sulfamethazine reduces divergence to 0.0031722794314617488
Removing query tranquilizer reduces divergence to 0.003171322366645038
Removing query animal psychology reduces divergence to 0.0031709345772320202
No more queries to remove
We can see that the algorithm was able to find a set of queries with a KL divergence of 0.0031, a value 10 times lower than the initial KL divergence of 0.03.
Looking into the logs, we can also see that the removed queries are mostly those that are very specific to a single faculty (especially medical terms). This is in line with our intuition - the queries that are too specific to a single faculty are likely to cause a high KL divergence from the global distribution.
pd.DataFrame({
'totals': totals['prob'],
'selection': get_series_for_queries(best_query_set),
}).sort_values('totals', ascending=False).plot(kind='bar', figsize=(20, 10))
<Axes: xlabel='faculty'>
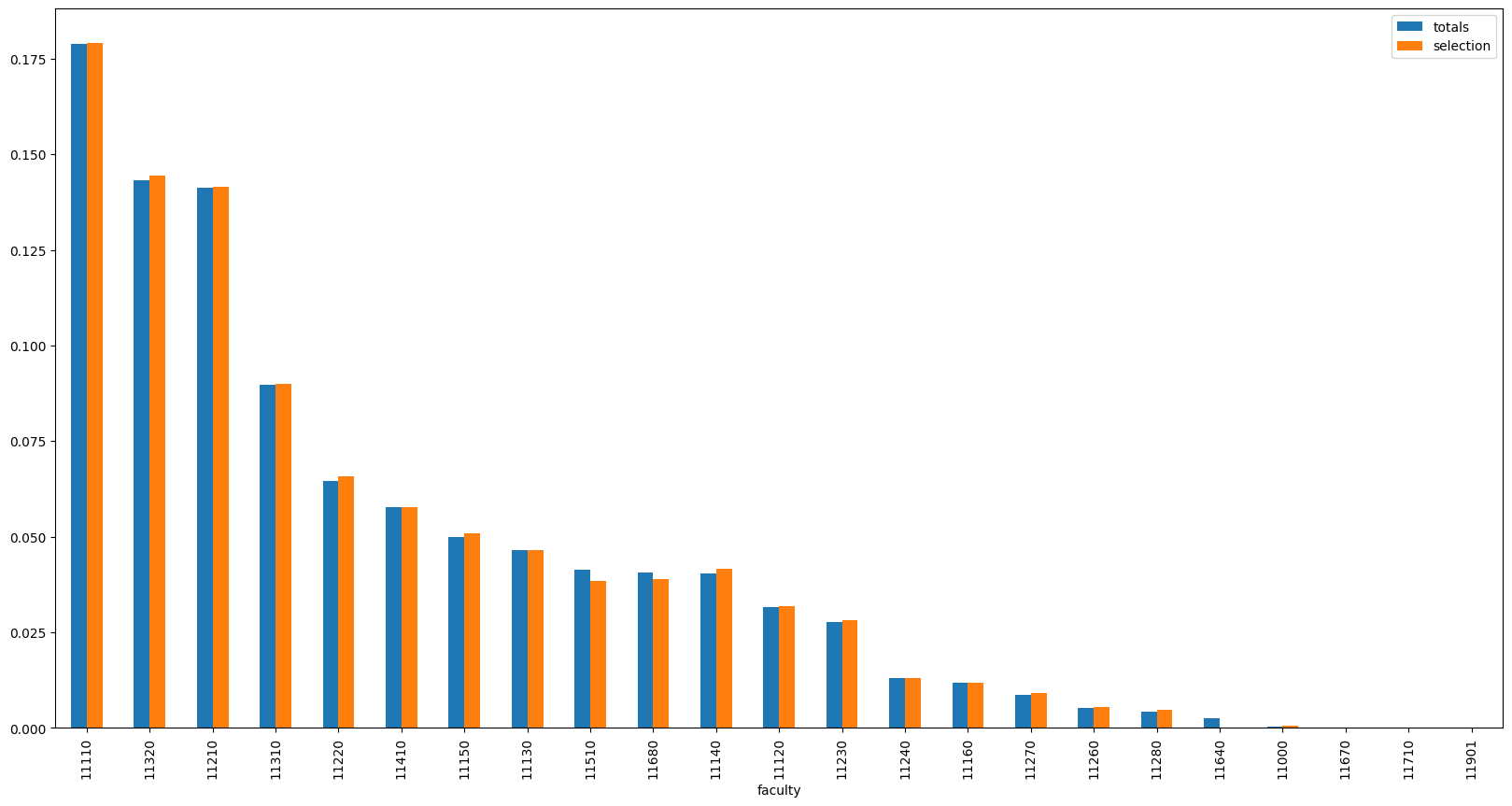
We can now store the optimized set of queries and use it for the benchmarking.
pd.Series(best_query_set).to_csv('best_queries.csv')
import pandas as pd
# Now that we have the best queries, we can retrieve the search results with their respective positions (0-30)
# Because we've removed the no-faculty publications, we need to reload the search results dataframe from the stored file.
best_query_set = pd.read_csv('best_queries.csv')['0'].tolist()
df = pd.read_csv('./search_results.csv');
After loading, we can now filter the search results to only include the best queries.
df[df['query'].isin(best_query_set)]
| Unnamed: 0 | id | year | name | faculty | faculty_name | query | |
|---|---|---|---|---|---|---|---|
| 30 | 0 | 140800 | 2011.0 | Basics of finance | -1 | Unknown faculty | finance |
| 31 | 1 | 117899 | 2010.0 | On Municipalities and their Finances | 11210 | Faculty of Arts | finance |
| 32 | 2 | 145860 | 2011.0 | Personal Finance | 11210 | Faculty of Arts | finance |
| 33 | 3 | 38694 | 2009.0 | Finance and Financial Activity of the State | 11220 | Faculty of Law | finance |
| 34 | 4 | 31340 | 2009.0 | The Reform of Public Finance in the Czech Repu... | 11220 | Faculty of Law | finance |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 12509 | 25 | 287421 | 2014.0 | Within-Day and Weekly Variations of Thrombolys... | 11140 | Faculty of Medicine in Pilsen | thrombolytic |
| 12510 | 26 | 594262 | 2021.0 | Stroke 20 20: Implementation goals for intrave... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
| 12511 | 27 | 610284 | 2022.0 | Stroke thrombectomy (+- thrombolysis), and not... | 11120 | Third Faculty of Medicine | thrombolytic |
| 12512 | 28 | 479107 | 2011.0 | How accurately can the aetiology of cardiac ar... | 11680 | Central Library of Charles University | thrombolytic |
| 12513 | 29 | 113007 | 2010.0 | Endovascular Recanalization in the Treatment o... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
3634 rows × 7 columns
We can see that the filtered data indeed contains results from various faculties.
On top of this, we also have the result ranking for each query in the Unnamed: 0 column. We can use this data in the DCG calculation.
df.rename(columns={'Unnamed: 0': 'position'})
| position | id | year | name | faculty | faculty_name | query | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 612484 | 2022.0 | Statins and liver | 11110 | First Faculty of Medicine | statin |
| 1 | 1 | 335691 | 2013.0 | Statins and myopathy | 11140 | Faculty of Medicine in Pilsen | statin |
| 2 | 2 | 594199 | 2016.0 | Do we use statins enough? News in the combinat... | 11140 | Faculty of Medicine in Pilsen | statin |
| 3 | 3 | 603996 | 2021.0 | Statins and COVID-19: Are they each other indi... | 11110 | First Faculty of Medicine | statin |
| 4 | 4 | 503698 | 2015.0 | Statins | 11110 | First Faculty of Medicine | statin |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 12509 | 25 | 287421 | 2014.0 | Within-Day and Weekly Variations of Thrombolys... | 11140 | Faculty of Medicine in Pilsen | thrombolytic |
| 12510 | 26 | 594262 | 2021.0 | Stroke 20 20: Implementation goals for intrave... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
| 12511 | 27 | 610284 | 2022.0 | Stroke thrombectomy (+- thrombolysis), and not... | 11120 | Third Faculty of Medicine | thrombolytic |
| 12512 | 28 | 479107 | 2011.0 | How accurately can the aetiology of cardiac ar... | 11680 | Central Library of Charles University | thrombolytic |
| 12513 | 29 | 113007 | 2010.0 | Endovascular Recanalization in the Treatment o... | 11150 | Faculty of Medicine in Hradec Králové | thrombolytic |
12514 rows × 7 columns